install.packages("tidyverse")Reproducible Data Analysis Day 4: Data loading and tidying with tidyverse
Learning Objectives
By the end of this class, you should
- Understand what packages are and how to use them
- Be able to load data into R
- Be able to use the six main data frame manipulation functions (and pipes) in
dplyr
What is data wrangling?
Manipulation of data frames means many things to many researchers: we often select certain observations (rows) or variables (columns), we often group the data by a certain variable(s), or we even calculate summary statistics. This is collectively referred to as “Data Wrangling” (to “wrangle” means to organize something unruly, like livestock).
Create the project
As before, create a new project to practice today’s code in the data-analysis-course folder on your Desktop. Call it gapminder-analysis, since we will be analyzing a dataset called “gapminder” (we will continue to use this dataset for the remainder of the course). Also, create a folder called data_raw inside the project.
Today we will be loading data from an external file (gapminder.csv). Download the file from this link, and put it in the data_raw folder in your project: https://www.dropbox.com/s/fdirlsnxlzy53qq/gapminder.csv?dl=0
Installing and loading packages
For this lesson, we will use functions that are not included in R by default (“Base R”). Packages are collections of code that expand R’s functionality. There are nearly 200,000 packages available on CRAN, the biggest repository of R packages.
To install a new package, use the install.packages() function, specifying the name of the package in quotation marks. Here, we will install a package called tidyverse:
Actually the “tidyverse” is a collection of R packages designed for data science, including readr(for loading data), dplyr (for data manipulation), ggplot (for plotting), and others. These packages are specifically designed to work harmoniously together. Some of these packages will be covered along this course, but you can find more complete information here: https://www.tidyverse.org/.
You only have to install the package once, which downloads it to your computer. But each time you want to use the package in an R session, you need to load it with the library() function, like this:
library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.2 ✔ tibble 3.3.0
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.1.0
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsNotice that since tidyverse contains multiple packages, it loads each of those and prints a message for each.
Somewhat confusingly, you don’t need to put the name of the pacakge quotation marks when using library(), but you do when using install.packages(). Also, you can only load one package at a time using library(), so if you needed to load multiple packages, you would need to do it like this (this example shows two packages included in tidyverse):
library(dplyr)
library(ggplot)Loading data
We can use the read_csv() function from the readr package included in tidyverse to load the data into R.
gapminder <- read_csv("data_raw/gapminder.csv")Rows: 1704 Columns: 6
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (2): country, continent
dbl (4): year, lifeExp, pop, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.R loads the data as a dataframe, also called a “tibble”. But what is a dataframe?
What are dataframes?
When we loaded the data into R, it got stored as an object of class tibble, which is a special kind of dataframe (the difference is not important for our purposes, but you can learn more about tibbles here). Dataframes are the de facto data structure for most tabular data, and what we use for statistics and plotting. Data frames can be created by hand, but most commonly they are generated by functions like read_csv(); in other words, when importing spreadsheets from your hard drive or the web.
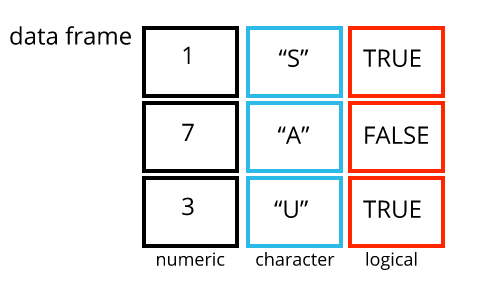
A dataframe is the representation of data in the format of a table where the columns are vectors that all have the same length. Because columns are vectors, each column must contain a single type of data (e.g., characters, integers, factors). For example, here is a figure depicting a dataframe comprising a numeric, a character, and a logical vector.
Inspect the data
You can see the contents of a dataframe by typing its name in the R console:
gapminder# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rowsR gives you some useful summary information: the number of rows (1,704) and columns (6) and the type of each column (chr is character, dbl is numeric).
About the “gapminder” dataset
This dataset includes economic statistics from various countries over time, from https://gapminder.org.
The meaning of some columns is obvious (country, continent, year), but not others. Here is a short explanation of those:
pop: PopulationlifeExp: Life expectancy (寿命)gdpPercap: GDP per capita (一人当たりの国内総生産)
Sort data with arrange()
First provide the name of the dataframe, then the column to sort by:
arrange(gapminder, lifeExp)# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Rwanda Africa 1992 23.6 7290203 737.
2 Afghanistan Asia 1952 28.8 8425333 779.
3 Gambia Africa 1952 30 284320 485.
4 Angola Africa 1952 30.0 4232095 3521.
5 Sierra Leone Africa 1952 30.3 2143249 880.
6 Afghanistan Asia 1957 30.3 9240934 821.
7 Cambodia Asia 1977 31.2 6978607 525.
8 Mozambique Africa 1952 31.3 6446316 469.
9 Sierra Leone Africa 1957 31.6 2295678 1004.
10 Burkina Faso Africa 1952 32.0 4469979 543.
# ℹ 1,694 more rowsThe default setting is to sort from small to large. To sort in the reverse (descending) direction, use desc():
arrange(gapminder, desc(lifeExp))# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Japan Asia 2007 82.6 127467972 31656.
2 Hong Kong, China Asia 2007 82.2 6980412 39725.
3 Japan Asia 2002 82 127065841 28605.
4 Iceland Europe 2007 81.8 301931 36181.
5 Switzerland Europe 2007 81.7 7554661 37506.
6 Hong Kong, China Asia 2002 81.5 6762476 30209.
7 Australia Oceania 2007 81.2 20434176 34435.
8 Spain Europe 2007 80.9 40448191 28821.
9 Sweden Europe 2007 80.9 9031088 33860.
10 Israel Asia 2007 80.7 6426679 25523.
# ℹ 1,694 more rowsYou can sort on multiple columns. Ties will be sorted by the next column:
arrange(gapminder, continent, lifeExp)# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Rwanda Africa 1992 23.6 7290203 737.
2 Gambia Africa 1952 30 284320 485.
3 Angola Africa 1952 30.0 4232095 3521.
4 Sierra Leone Africa 1952 30.3 2143249 880.
5 Mozambique Africa 1952 31.3 6446316 469.
6 Sierra Leone Africa 1957 31.6 2295678 1004.
7 Burkina Faso Africa 1952 32.0 4469979 543.
8 Angola Africa 1957 32.0 4561361 3828.
9 Gambia Africa 1957 32.1 323150 521.
10 Guinea-Bissau Africa 1952 32.5 580653 300.
# ℹ 1,694 more rowsNarrow down columns with select()
First provide the name of the dataframe, then the columns to select:
select(gapminder, year, country, gdpPercap)# A tibble: 1,704 × 3
year country gdpPercap
<dbl> <chr> <dbl>
1 1952 Afghanistan 779.
2 1957 Afghanistan 821.
3 1962 Afghanistan 853.
4 1967 Afghanistan 836.
5 1972 Afghanistan 740.
6 1977 Afghanistan 786.
7 1982 Afghanistan 978.
8 1987 Afghanistan 852.
9 1992 Afghanistan 649.
10 1997 Afghanistan 635.
# ℹ 1,694 more rowsSaving your output
Notice that although we have used several functions, gapminder is still the same:
gapminder# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rowsThis is because we have not saved any of the output. To do that, you need to create a new object with <-. You can call the object whatever you want, but use a name that is easy to remember:
gapminder_gdp <- select(gapminder, year, country, gdpPercap)About pipes
During data analysis, we often need to perform several intermediate steps. It is not convenient to save the output of each since you have to think of names for each output object, and may not even use them for anything later. A better way to do this is using something called the “pipe”. The pipe is written like this: |> (some packages also write it like this: %>%).
The pipe takes the output from one function and passes it to the input of the next function. You can think of it as saying “and then”:
- Do this and then do this, and then do this…
- Do this
|>do this,|>do this…
We can even use the pipe just with one function. Read the following as “start with gapminder and then select only year, country, and population”:
gapminder |> select(year, country, pop)# A tibble: 1,704 × 3
year country pop
<dbl> <chr> <dbl>
1 1952 Afghanistan 8425333
2 1957 Afghanistan 9240934
3 1962 Afghanistan 10267083
4 1967 Afghanistan 11537966
5 1972 Afghanistan 13079460
6 1977 Afghanistan 14880372
7 1982 Afghanistan 12881816
8 1987 Afghanistan 13867957
9 1992 Afghanistan 16317921
10 1997 Afghanistan 22227415
# ℹ 1,694 more rowsThis becomes very useful when we want to do multiple steps. Read this as “start with gapminder, and then select only year, country, and population, and then arrange by year”:
gapminder |>
select(year, country, pop) |>
arrange(year)# A tibble: 1,704 × 3
year country pop
<dbl> <chr> <dbl>
1 1952 Afghanistan 8425333
2 1952 Albania 1282697
3 1952 Algeria 9279525
4 1952 Angola 4232095
5 1952 Argentina 17876956
6 1952 Australia 8691212
7 1952 Austria 6927772
8 1952 Bahrain 120447
9 1952 Bangladesh 46886859
10 1952 Belgium 8730405
# ℹ 1,694 more rowsWe can make it easier to read by putting each step on its own line (the result is exactly the same, since R ignores spaces and line breaks):
gapminder |>
select(year, country, pop) |>
arrange(year)# A tibble: 1,704 × 3
year country pop
<dbl> <chr> <dbl>
1 1952 Afghanistan 8425333
2 1952 Albania 1282697
3 1952 Algeria 9279525
4 1952 Angola 4232095
5 1952 Argentina 17876956
6 1952 Australia 8691212
7 1952 Austria 6927772
8 1952 Bahrain 120447
9 1952 Bangladesh 46886859
10 1952 Belgium 8730405
# ℹ 1,694 more rowsPipes are very useful because you don’t have to save each intermediate step. This is a very useful way to manipulate data. Therefore, the rest of the lesson will use the pipe, to help you get used to it.
Subset rows with filter()
Use the filter() function to only keep rows that meet a certain condition. For example, let’s only keep the data in Europe:
gapminder |> filter(continent == "Europe")# A tibble: 360 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Albania Europe 1952 55.2 1282697 1601.
2 Albania Europe 1957 59.3 1476505 1942.
3 Albania Europe 1962 64.8 1728137 2313.
4 Albania Europe 1967 66.2 1984060 2760.
5 Albania Europe 1972 67.7 2263554 3313.
6 Albania Europe 1977 68.9 2509048 3533.
7 Albania Europe 1982 70.4 2780097 3631.
8 Albania Europe 1987 72 3075321 3739.
9 Albania Europe 1992 71.6 3326498 2497.
10 Albania Europe 1997 73.0 3428038 3193.
# ℹ 350 more rowsModify data with mutate()
For example, we could change the units of population to millions of people:
gapminder |> mutate(pop = pop / 1000000)# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8.43 779.
2 Afghanistan Asia 1957 30.3 9.24 821.
3 Afghanistan Asia 1962 32.0 10.3 853.
4 Afghanistan Asia 1967 34.0 11.5 836.
5 Afghanistan Asia 1972 36.1 13.1 740.
6 Afghanistan Asia 1977 38.4 14.9 786.
7 Afghanistan Asia 1982 39.9 12.9 978.
8 Afghanistan Asia 1987 40.8 13.9 852.
9 Afghanistan Asia 1992 41.7 16.3 649.
10 Afghanistan Asia 1997 41.8 22.2 635.
# ℹ 1,694 more rowsIf we provide a new column name, that column will be added
gapminder |> mutate(pop_mil = pop / 1000000)# A tibble: 1,704 × 7
country continent year lifeExp pop gdpPercap pop_mil
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779. 8.43
2 Afghanistan Asia 1957 30.3 9240934 821. 9.24
3 Afghanistan Asia 1962 32.0 10267083 853. 10.3
4 Afghanistan Asia 1967 34.0 11537966 836. 11.5
5 Afghanistan Asia 1972 36.1 13079460 740. 13.1
6 Afghanistan Asia 1977 38.4 14880372 786. 14.9
7 Afghanistan Asia 1982 39.9 12881816 978. 12.9
8 Afghanistan Asia 1987 40.8 13867957 852. 13.9
9 Afghanistan Asia 1992 41.7 16317921 649. 16.3
10 Afghanistan Asia 1997 41.8 22227415 635. 22.2
# ℹ 1,694 more rowsCalculate summary statistics with summarize()
For example, let’s calculate the overall mean population:
gapminder |> summarize(mean_pop = mean(pop))# A tibble: 1 × 1
mean_pop
<dbl>
1 29601212.As another example, let’s calculate the total population over all the data:
gapminder |> summarize(total_pop = sum(pop))# A tibble: 1 × 1
total_pop
<dbl>
1 50440465801Use group_by() to do calculations per group
However, it is often more useful to calculate such summary statistics for particular groups. To do this, first specify the groups with group_by():
gapminder |> group_by(continent)# A tibble: 1,704 × 6
# Groups: continent [5]
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rowsNext, use summarize() to calculate the summary statistic:
gapminder |>
group_by(continent) |>
summarize(mean_pop = mean(pop))# A tibble: 5 × 2
continent mean_pop
<chr> <dbl>
1 Africa 9916003.
2 Americas 24504795.
3 Asia 77038722.
4 Europe 17169765.
5 Oceania 8874672.summarize() and mutate() are related since they both create new columns (or over-write existing ones). You can remember the key difference between them is that mutate() keeps the number of rows the same, whereas summarize() always decreases the number of rows.
Summary
Here is a list of the data-wrangling commands we have learned so far:
- Sort data with
arrange() - Narrow down columns with
select() - Filter rows with
filter() - Modify data with
mutate() - Summarize data with
summarize() - Group data with
group_by()
By combining these functions, each of which is fairly simple, with the pipe (|>), you can construct sophisticated data analysis pipelines.
Submitting the homework
Go to Moodle, click on the Day 4 Homework assignment, and click on the link to accept the assignment. This will create the repo in your GitHub account.
Next, clone the remote repo to your local machine. Then, edit the file day_04_homework.R file in RStudio. Make sure that your code runs without errors. Once you have done so, commit your changes and push to the remote. Don’t forget to push! If you don’t push, your work will not be submitted.
You can double check that you’ve successfully submitted (pushed) the homework by visiting your GitHub repo in your browser (use the same link in Moodle that you clicked to accept the assignment). Make sure the homework file there shows the same changes that you made in RStudio.
Be sure to do this BY THE DEADLINE, or your work will not be counted.
Attributions
These materials were modified by Joel H. Nitta from those posted at https://swcarpentry.github.io/r-novice-gapminder/ and https://datacarpentry.org/R-ecology-lesson under the Creative Commons Attribution (CC BY 4.0) license.